Ever wanted to leverage the geometry of data for boosting Gaussian Processes learning performances without getting into complex differential geometry and manifolds stuff?
Our last NeurIPS paper “Implicit Manifold Gaussian Process Regression” will help you in achieving that!
Read it here!
In this work we propose a Gaussian Process (GP) regression technique capable of learning a given function on non-Euclidean domain while inferring the implicit manifold structure directly from data in a fully differentiable way.
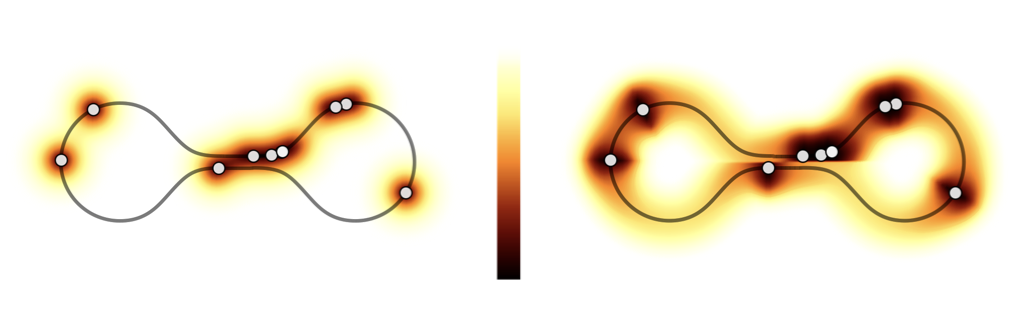
GP on Manifolds leverages geometric kernels to account for the space geometry. Such kernels are built through the eigenvalues and eigenfunctions of the Laplace-Beltrami on the given manifold. Such eigenvalues and eigenfunction can be computed analytically or numerically via mesh-based methods whenever the knowledge of the underlying manifold is available. What can we do in those case where the manifold geometry is not available?
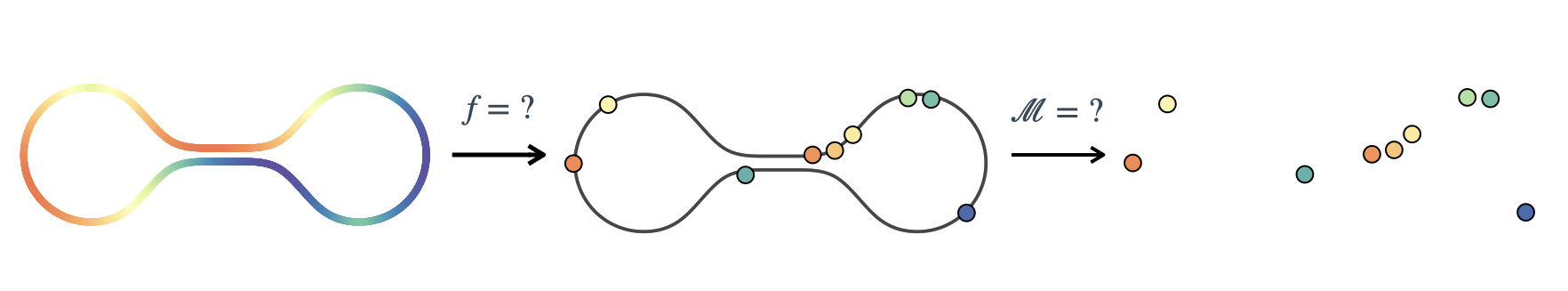 Consider the problem of learning the function $f : \mathcal{M} \rightarrow \mathbb{R}$ on Riemannian manifolds, $\mathcal{M}$. We target the scenario where the manifold, $\mathcal{M}$, is unknown. We want to infer both $f$ and $\mathcal{M}$ from the available data.
Consider the problem of learning the function $f : \mathcal{M} \rightarrow \mathbb{R}$ on Riemannian manifolds, $\mathcal{M}$. We target the scenario where the manifold, $\mathcal{M}$, is unknown. We want to infer both $f$ and $\mathcal{M}$ from the available data.
Our approach operates in both supervised and semi-supervised learning scenario. In the latter case both labeled and unlabeled data are leveraged in order to infer the correct underlying geometry. In order perform GP on manifolds, it is necessary to approximate the Laplace-Beltrami operator. This, in turn, will provide the eigenvalues and the eigenfunctions necessary to recover the geometric kernel.
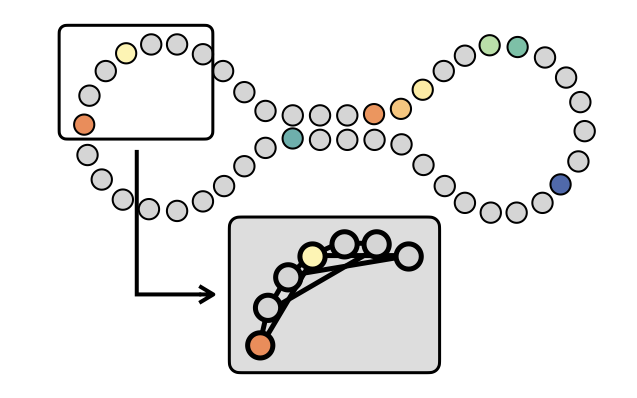 In order to achieve that, we construct a graph-based Laplacian on top the available datapoints. Two key points: 1) we build the graph via symmetric KNN so that our Laplacian results in a sparse matrix; 2) we construct an appropriate re-normalized random-walk Laplacian that guarantees point wise convergence to the Laplace-Beltrami operator independently from the sampling density.
In order to achieve that, we construct a graph-based Laplacian on top the available datapoints. Two key points: 1) we build the graph via symmetric KNN so that our Laplacian results in a sparse matrix; 2) we construct an appropriate re-normalized random-walk Laplacian that guarantees point wise convergence to the Laplace-Beltrami operator independently from the sampling density.
Marginal likelihood optimization is performed on the graph without reconstructing an explicit representation of the kernel. Both graph and GP hyper parameters are optimized at the same time, linking the problem of learning the manifold with the one of learning on the manifold. Crucially, we focus on hyperparameters optimization of the Matern kernel, whose polynomial spectral density allows us to device a sparse, matrix-free and fully differentiable optimization strategy.
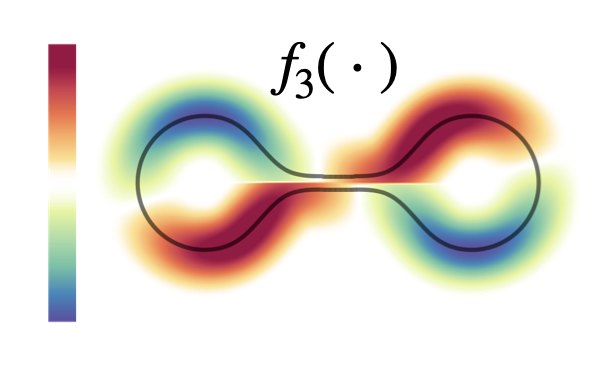 In order to maintain high performance at query-time we reconstruct continuous eigenfunctions via a KNN-based Nystrom formula.
In order to maintain high performance at query-time we reconstruct continuous eigenfunctions via a KNN-based Nystrom formula.
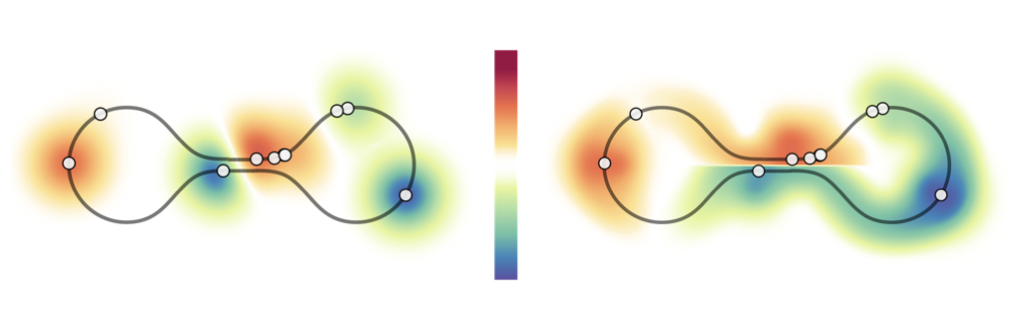
 The approximated geometric kernel accounts for the underlying manifold’s geometry.
The approximated geometric kernel accounts for the underlying manifold’s geometry.
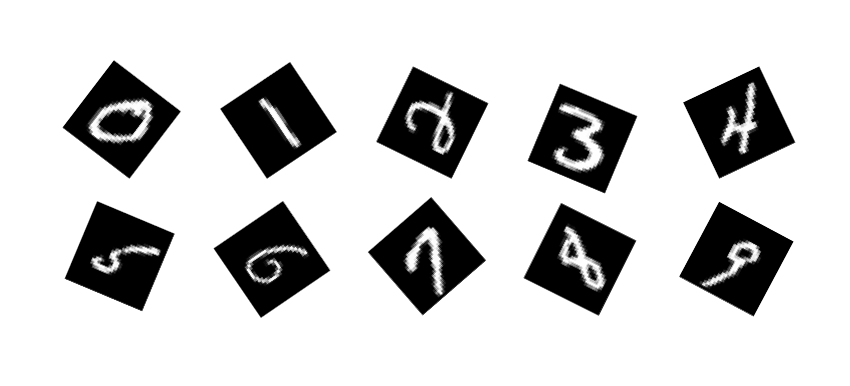 Thanks to its sparse structure, our method scales up hundreds of thousand of data point and can be applied to any high dimensional dataset where the manifold hypothesis holds.
Thanks to its sparse structure, our method scales up hundreds of thousand of data point and can be applied to any high dimensional dataset where the manifold hypothesis holds.
If you are interested in using our work, head over to our repo on GitHub. The jupyter notebook examples will guide you through!